POSTS
Sliding - What matters?
We have previously measured the sliding distance of LacI on DNA in vivo and concluded that the transcription factor uses a combination of 3D diffusion in the cytoplasm and 1D diffusion on the DNA to speed up the search for the operator. We now dig deeper into the sliding mechanism to figure out more about how the TF slides on DNA. How fast is the sliding? Is it smooth or does it include frequent microscopic jumps (hopping)? What is the influence of multiple DNA binding domains allowing the TF to transfer between DNA strands and what is the effect of molecular crowding? Using Monte Carlo Simulations scheme to realize the search process for one or two operator sites, we try to define a physically reasonable sets of parameters for which the model can explain the observed in vivo measurements.
We find that non-specific binding to DNA is improbable at first contact and that the sliding LacI protein binds at high probability when reaching the operator. We also conclude that the contribution of hopping to the overall search speed is negligible although physically unavoidable and we see an unexpectedly high 1D diffusion constant on non-specific DNA sequences. Including a mechanism of inter-segment transfer between distant DNA segments does not bring down the 1D diffusion to the expected fraction of the in vitro value, suggesting a more complicated explanation than molecular crowding.
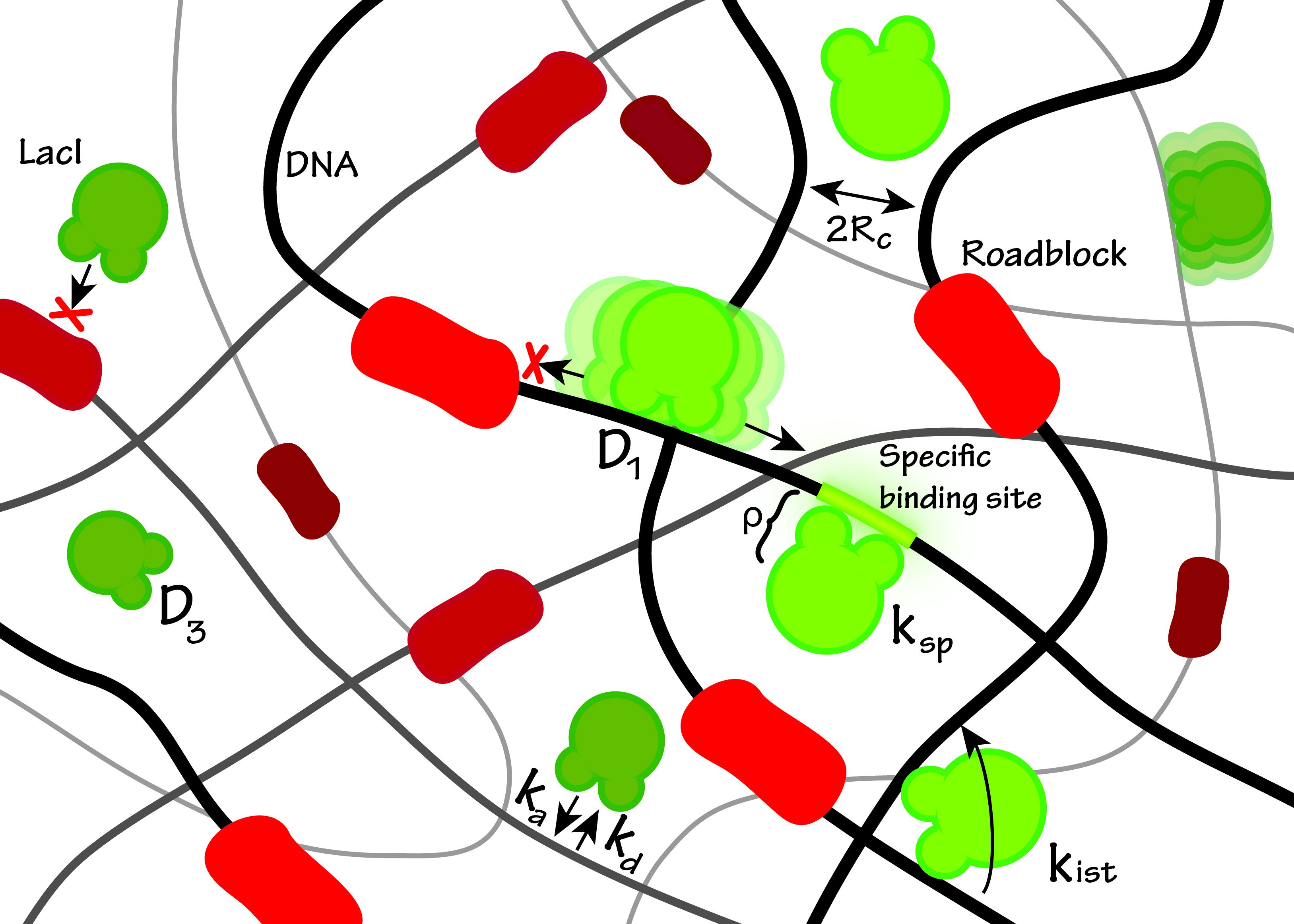
Cartoon of the search process and the fundamental parameters involved in our model. Following dissociation the searching protein (green) can associate with rate ka to a nonspecific stretch of DNA (black) given that no other proteins (red) are in the way. Given a successful nonspecific association the protein may diffuse in 1D along the length of the DNA or perform intersegment transfer with rate kIST.
Check out the full publication in NAR