Accessing Genetic Information in the Living Cell
In this project we will combine high-resolution single-molecule investigations in vitro with molecular dynamics (MD) simulations and experiments on sequence-specific search kinetics in living cells, in order to decipher the regulatory genetic codes.
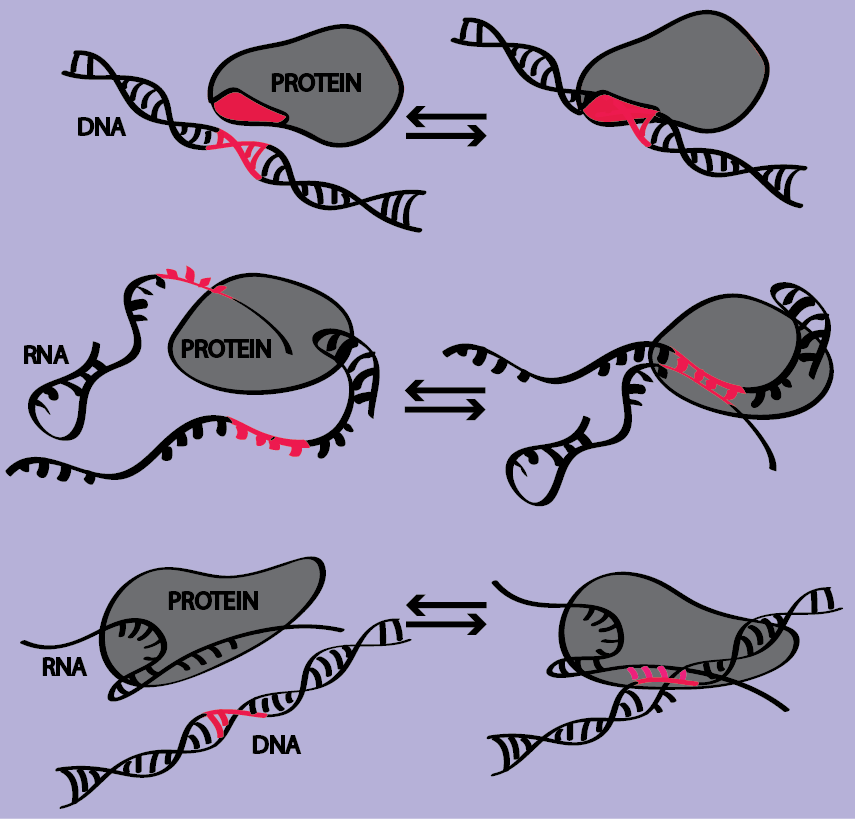
Our understanding of the genetic code is essentially limited to the protein coding regions, for which the rules of decoding triplet nucleotides by tRNA adaptors are well known. However, hidden in the code are also instructions for how RNA and protein expression are dynamically regulated in space and time. This information is as important for life as the mechanistic function of the individual molecules. The codes are read by regulatory molecules that provide access to the correct genetic information at the right time. The average interaction patterns are often known from genome-wide, sequencing-based methods, but very little is known about how the specific genetic information is quickly and accurately searched for and retrieved when needed.
The fact that DNA has a uniform structure built from a few recurring building blocks linked into a linear sequence, facilitates its maintenance and replication. However, it also presents all macromolecules that need to retrieve genetic information with a substantial challenge; how do you find the right combination of bases in the vast pool of very similar sequences?
Cells have evolved two radically different search strategies in order to scan a genome or transcriptome in search of a specific piece of genetic code. Some proteins, e.g. transcription factors or nucleosomes, recognize a specific or preferred DNA sequence through interactions in the grooves, whilst other macromolecules are dynamically programmed by RNA molecules to recognize complementary nucleic acid sequences. Examples of the latter are Argonaute and Hfq programmed by interference RNA and small RNA, respectively, searching for complementary target mRNAs, CRISPR/Cas9 programmed by guide RNA to interrogate double-stranded DNA and RecA programmed by ssDNA searching for the other allele in homologous recombination.


The project is an interdisciplinary effort including principal investigators at Uppsala University, Stockholm University and KTH. The project is funded by reserach grants from the Knut and Alice Wallenberg foundation and The Swedish Research Council.
The Knut and Alice Wallenberg Foundation
If you have general questions about the project, please contact: irmeli.barkefors@icm.uu.se







