POSTS
Better Analysis of Noisy Multistate Diffusion Trajectories
Single particle tracking offers a non-invasive high-resolution probe of biomolecular reactions inside living cells. However, efficient data analysis methods that correctly account for various noise sources are needed to realize the full quantitative potential of the method. In a recent publication we report a new algorithms for hidden Markov-based analysis of single particle tracking data, which incorporate most sources of experimental noise, including heterogeneous localization errors and missing positions.
Compared to previous implementations, the algorithms offer significant speed-ups, support for a wider range of inference methods, and a simple user interface. This will enable more advanced and exploratory quantitative analysis of single particle tracking data.
Read the paper in Biophysical Journal.
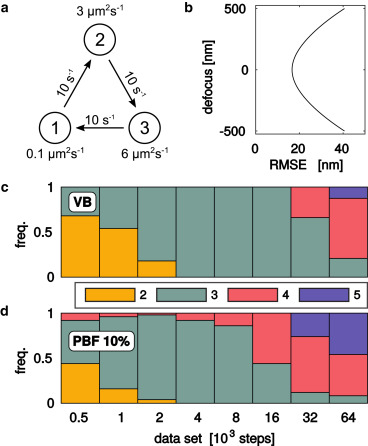
Figure: Statistical model selection. We generated a range of synthetic data sets with (a) three diffusive states and (b) defocus-dependent localization root-mean-square errors (RMSE), and estimated the number of states using © variational maximum evidence (VB) and (d) cross-validation using variational pseudo-Bayes factors (CV) and 10% of the data in the validation set.