POSTS
The right drug for the right bug
Most of us take functioning antibiotics for granted; it is hard to imagine a world where death by pneumonia, routine surgery, or birth complications is commonplace. Yet that might very well be the reality in the not-too-distant future if the resistance crisis is not acknowledged and mitigated. Globally, we are using more and more antibiotics and while generally effective to cure bacterial diseases, the increased use of these drugs creates a perfect breeding ground for resistance mutation. In other words, the bacteria are evolving to cope with the antibiotics, slowly rendering them ineffective. Using antibiotics in moderation is one of the prescribed cures.
We have previously developed a test to measure antibiotic resistance in urinary tract infections. The test tells you know if the patient’s problem is caused by bacteria and what particular antibiotic would take them out. But for complex conditions with unknown species, like sepsis , our only option right now is to hit blindly, using precious last-resort drugs.
To make a resistance test for complex infections, the identity of the culpable bacteria must be known, which takes around 24 hours with current protocols. But many sepsis patients do not have that kind of time. For every hour the patient is left untreated, the risk of a fatal outcome increases dramatically.
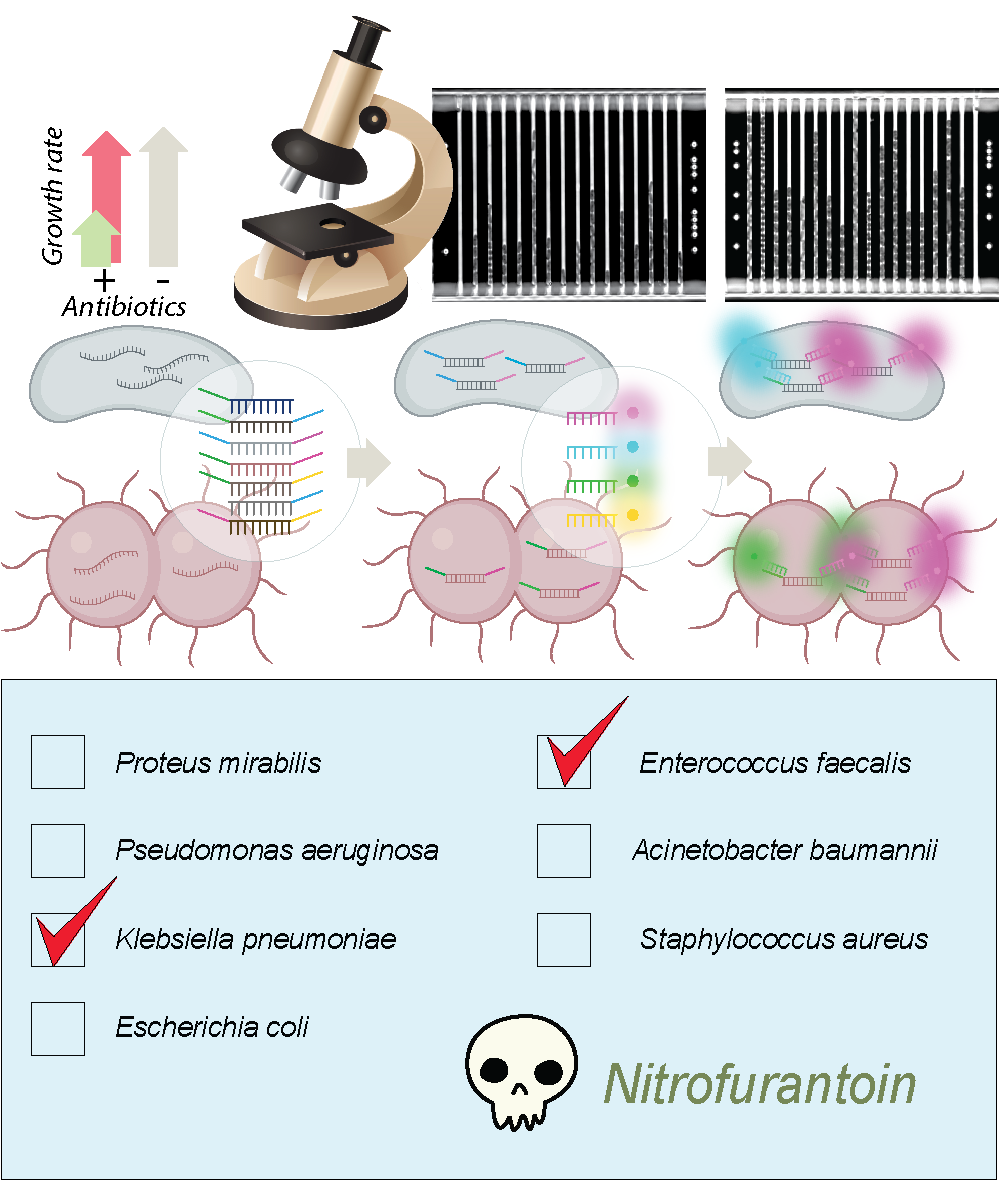
We have now developed our protocol to include the sought-after bacterial identification step. We load the sample into a microfluidic culture chip that works a bit like a microscopic sieve, capturing the bacteria in thousands of tiny culture channels. In the chip, the cells are exposed to different antibiotics, and we monitor their growth in real-time by an image analysis algorithm that compares with untreated cells and calculates if the drugs are effective or not. If the bacteria keeps growing in the presence of a certain antibiotic, chances are high that they are resistant to that particular drug.
After the growth-rate measurements have been made, we flood the chip with colored molecular probes targeting most of the common bacterial pathogens. Since the algorithm knows what color combination corresponds to which bacteria, and keeps track of the growth rate in each channel, it can match resistance and bacterial species and suggest the best course of treatment.
Read more about the method in Nature Communications