POSTS
Binding long or often - the secret of fast TF search
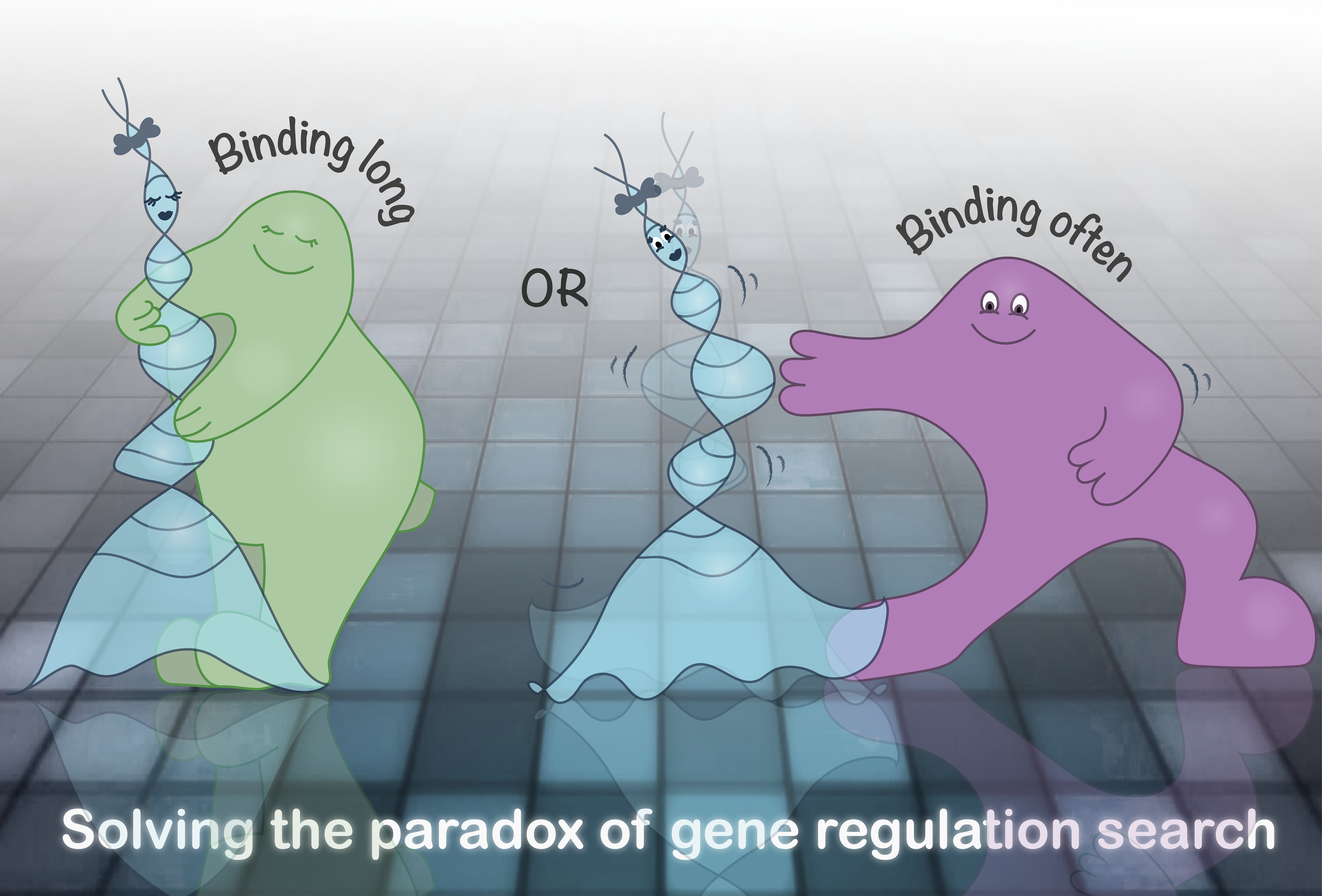
When things are bound together in the macroscopic world, that’s usually accomplished by them not detaching. It is reasonable to assume that molecules operate in much the same way. Contradicting this intuitive premise, we shown that DNA-binding proteins abide by a completely different set of rules. The proteins often detach, but to their target DNA sequence, they find their way back, time and time again. Maybe, you might think, it doesn’t matter all that much - to bind a long time or to bind often - but the result might explain one of the great mysteries of gene regulation; how can a DNA-binding protein search the entire genome for its target sequence without getting held up on the way?
Read the complete article in Science
Over an organism’s lifetime, its genome changes very little. What does change, constantly, are which proteins the cell produces in response to damage, changes in the environment, or stages in the reproductive cycle. The protein production is regulated by DNA-binding proteins that have evolved the ability to turn different genes on or off. Because the environment can change quickly, rapid adaptation is key. The DNA-binding proteins must find the correct DNA code among millions of base pairs, and do so fast.
When DNA-binding proteins search the genetic code for their target sequence, they slide along the DNA helix to speed up the process. When they finally find the right spot, they stay there; the interaction with the “correct” sequence prevents them from sliding along. This mechanism has been widely accepted to describe the search process. It is an appealing hypothesis, yes, but it presents an annoying problem - the DNA code is full of “almost correct” sequences. If the time a protein resides on a particular DNA motif was determined by the sequence, the searching proteins would constantly linger on sequences that resembled their target.
If the textbook explanation was correct, the DNA-binding proteins would get stuck all the time off target. Gene regulation would be very ineffective, but we know from previous studies that this is not the case. Our favorite protein, LacI, finds its target sequence among 4.6 million base pairs in a matter of minutes.
In an attempt to resolve this paradox, we allowed the DNA-binding protein LacI to slide back and forth on thousands of different DNA sequences mounted on a microchip. We attached a fluorescent molecule to the LacI protein to make it possible to measure how fast LacI adhered to the different sequences and how quickly it was released. The result was striking. Contradicting previous assumptions, the DNA sequence had little effect on how long LacI remained bound to the DNA. However, it was much more likely that the sliding LacI was held up briefly when the sequence was similar to the target sequence. In other words, DNA-binding proteins often leave also the sequence they are intended to regulate, but at the target site, they all but always make a very short journey before finding their way back again. On the macroscopic time scale, this looks like a stable interaction.
Our result, that DNA-binding proteins bind often rather than protractedly, explains how LacI can slide on the DNA sequence in search of its target without getting held up unnecessarily. LacI regulates the uptake of lactose in bacteria, but is of course just an example. The hundreds of different transcription factors that regulate our own genes likely act according to a similar principle.