POSTS
Structural flexibility explains efficient search process
With our coarse-grained molecular model of different conformation of the lac repressor, we demonstrate how disordered regions assist both rapid exploration of the DNA and recognition of the specific site. This conformational agility explains how DNA-targeting proteins can be both fast and accurate.
When things change in our environment, our cells have to respond accordingly. If they fail to do so, we are doomed. The molecules responsible for conveying the message are known as transcription factors, TFs. They quickly locate the correct gene, often within minutes, among millions of DNA bases in order to turn it on or off. To speed up the search, TFs slide along the DNA, and although this mechanism reduces the dimensionality of the search from 3D to 1D, it also introduces a dilemma; how does the TF recognize the correct site without getting held up on partially similar sequences in the genome?
In a new study which we recently published in The Journal of Physical Chemistry B, we show that conformational changes in the protein and the DNA might hold the key to this riddle. One conformation allows the protein to slide quickly on the DNA and the other allows the TF to interact with the specific sequence. By performing simulations of the TF-DNA interaction during sliding as well as in the encounter complexes, we show how different TF conformations establish favorable specific and electrostatic interactions with different shapes of DNA and that the conformational changes in both molecules are coupled in recognition.
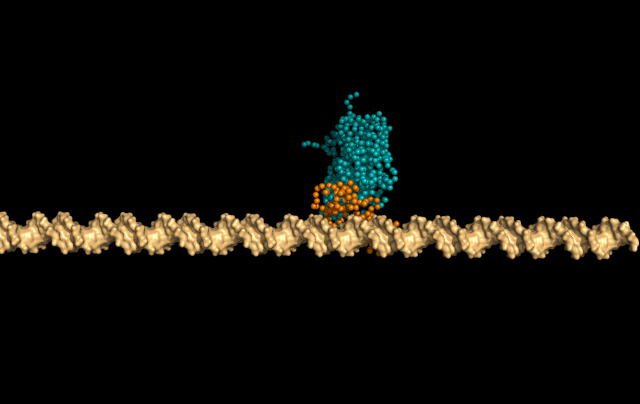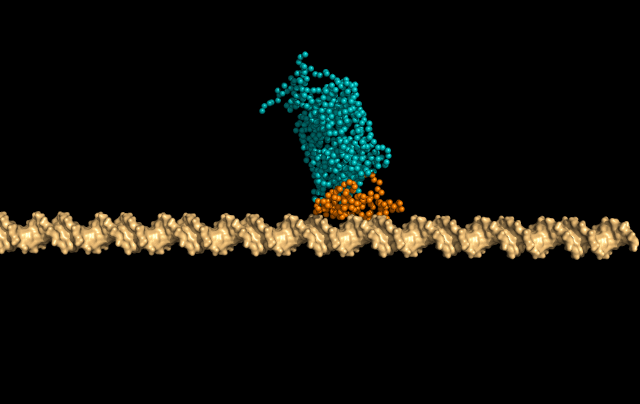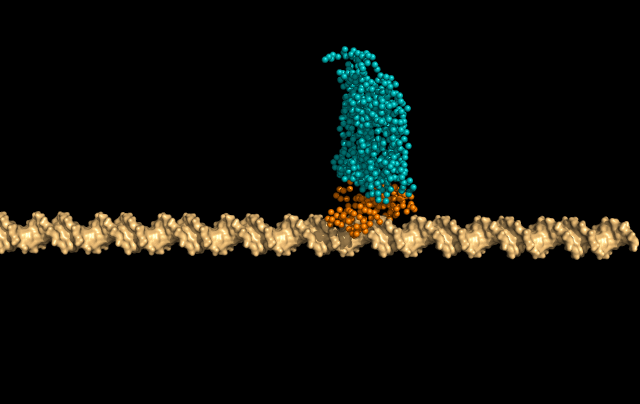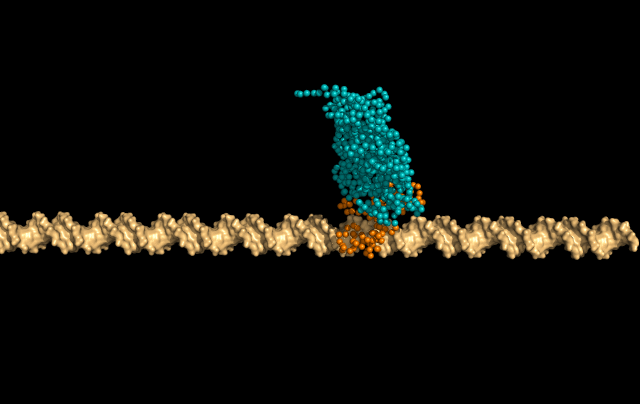
In this simulation, LacI is sliding faithfully in the DNA grooves, interrogating the DNA backbone in search of its target sequence.
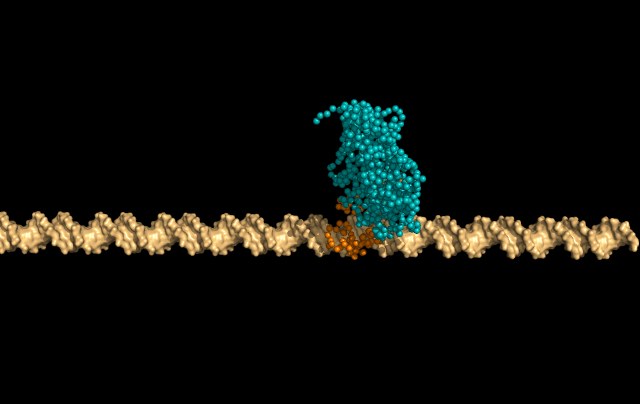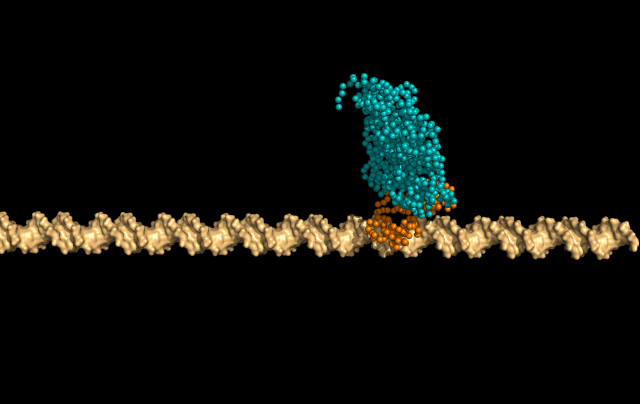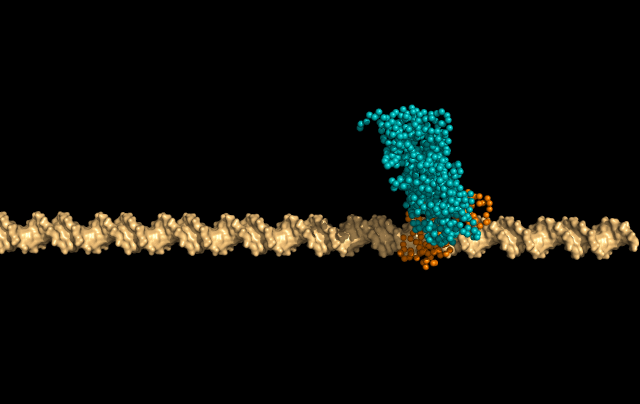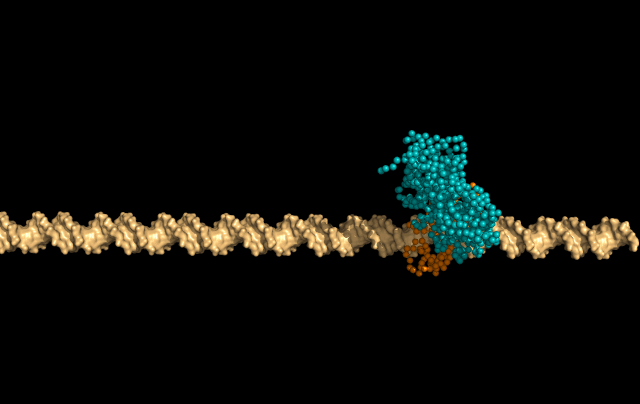
But the protein can also skip a DNA-turn or two, something we call hopping, to cover a larger distance of the helix faster.
One region of the TF, the hinge region, is particularly important for this conformational flexibility. If the hinge regions form helices at the protein-DNA interface, the TF interacts tightly with bent DNA, which is a characteristic of the specific complex; if the hinge region is disorganized, on the other hand, the TF is free to explore. Together with experimental results, these simulations show us how DNA-interacting proteins can be both fast and accurate in locating their target sequences in the genome. Read the paper