POSTS
MINFLUX - Resolving life
By following single molecules diffusing in the cell we can learn much about how biomolecules interact with each other in the cytoplasm. How the molecule switch between diffusive states can be used for example to estimate the reaction rate constants for molecules in equilibrium. One problem, however, has been that irreversible photo-bleaching greatly limits the individual trajectory length and thus the dynamic range of the reactions that can be studied. Historically, the only way to increase the trajectory length has been to reduce the light intensity and not collect so many photons per image, but as localization accuracy correlate negatively with the number of photons it has always been at the expense of the spatial resolution.
In collaboration with Stefan Hell’s group in Goettingen we have fundamentally changed the prerequisites for single molecule tracking in living cells. Instead of imaging the light from a molecule and then use the point spread function to appreciate its spatial location, we now follow the movement of the molecule using a hollow laser beam. The fluorescent molecule is trapped in the center of the laser beam where it is not excited and thus not emitting any photons. Only when the fluorophore deviates from the middle, we detect photons that we can then use to adjust the position of the laser and bring the molecule back to the center. Since the position of the molecule is defined indirectly by the laser position, we can use the available photons much more efficiently. In principle, one detected photon is enough to know that we are out of place, and only a few more are needed to calculate a new optimal position. This means that we can record trajectory lengths that are 100 times longer than what have been possible using standard techniques. The downside is that the system is complicated to set up since laser correction must occur on the microsecond scale.
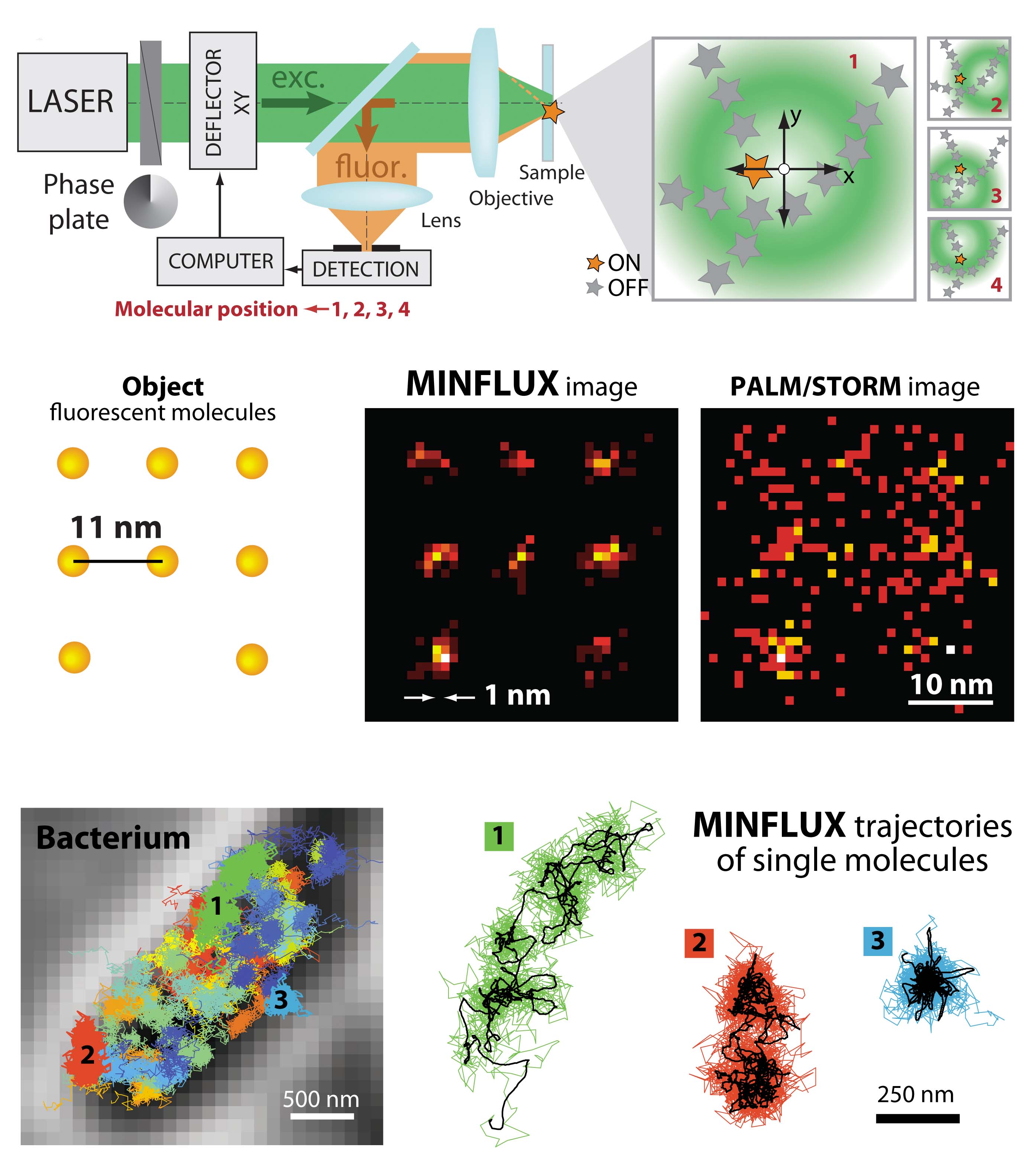
The new MINFLUX microscope will make it possible to finally study biological processes on the same spatial and temporal scale that can be reached by molecular dynamics simulations. The system that we have built in Uppsala will initially be used to study gene regulation and protein synthesis in living cells. An exciting journey into living matter at the molecular level has just begun!
Read the complete article in Science