POSTS
The small, the fast, and Shadowfax
The chemistry inside all living cells, from humans to bacteria, occurs at the nanometer length- and the microsecond time scale, a regime where scientists are frustratingly blind. Electron microscopes can help us see the supersmall, but this world is frozen. Fluorescence microscopy lets us follow the dynamics of intracellular processes, but the wavelength of light limits the spatial resolution to some hundred nanometers. A fraction of a micrometer might not sound like a lot, but this resolution is far from enough to see individual molecules. With clever optical tricks, we can improve the spatial resolution beyond this limit, but at the cost of time, something we cannot sacrifice when capturing fast chemical reactions.
Enter “Shadowfax”; like its mythological namesake, the Shadowfax microscope is swift as the wind. It can find and follow fast individual molecules inside living bacteria, but unfortunately for us and others interested in the chemistry of life, the actions of these molecules remain elusive.
One of the secrets behind the new technique is that it uses very few photons to localize the molecule. Instead of activating the fluorophore and recording an image from the positions of the emitted photons - which is the standard procedure for molecule tracking - the algorithm triangulates the location of a molecule by measuring the light emitted by the fluorescent molecule in a defined laser pattern. Since every time step consumes photons from a limited budget, rationing the photons means we can follow the molecules at a higher resolution. Another trick to maximize the speed is to reduce the number of equipment parts with mass that the system has to move frequently. Tracking both fast and slow molecules with good resolution is challenging, but with its large laser pattern, the system can keep track of the more elusive fast molecules.
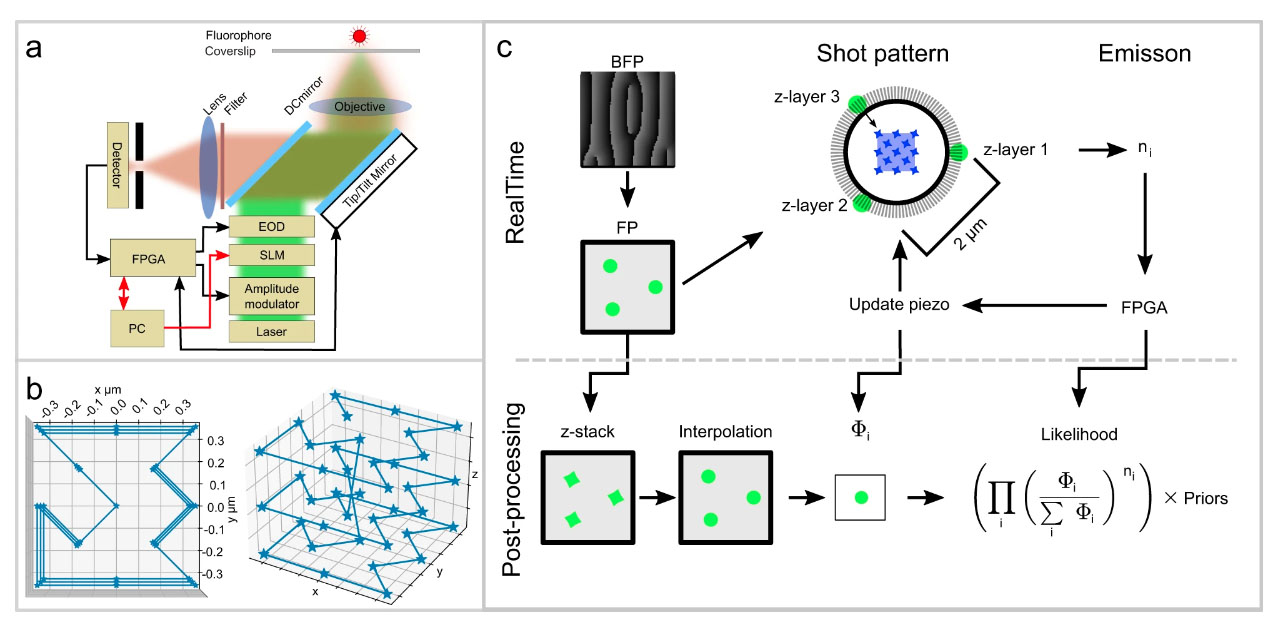
Before unleashing the microscope on biological molecules in living cells, we tested its performance carefully by tracking fluorescent beads moving on a stage. We could determine the position of the beads with 67 nm lateral and 109 nm axial precision and a time resolution of 0.84 ms; the measurements agreed with theoretical and simulated predictions.
Now that we’d confirmed the microscope could track fast-moving fluorophores with high precision, we needed a biological system to use as a proof-of-concept. Our choice fell on the E. coli trigger factor (TF). TF is biologically relevant as it helps newly translated proteins fold correctly by binding to the ribosome - or at least, that’s what we believe. We used Shadowfax to follow the movements of fluorescently labeled TF in living bacteria and analyzed the trajectories with a Hidden Markov Model to find the different binding states. We reasoned that free TF molecules diffuse faster than TF molecules bound to bulky ribosomes. To confirm what we saw was really ribosome binding, we used a mutated TF, unable to bind ribosomes, as a control. Although we reached sub-millisecond live-cell single-molecule tracking of TF and successfully detected different binding states, we were still hard-pressed to find the diffusive state transitions, e.i. more work needs to be done before we can determine the dynamics of intermolecular reactions at this time scale.
Read more in Nature Communications!